With a rapidly growing demand for high-quality animal protein, aquaculture is becoming the primary source of seafood for human diets. Clustered regularly interspaced short palindromic repeats (CRISPR) /Cas9 gene editing technology provides the most direct and powerful tool for genetic analysis and breeding of economic traits of important aquatic species because of its advantages of simple operation, wide selection of targets, low cost and high efficiency.
The oysters, as the representative bivalve mollusc, which are widely distributed and support major aquaculture and fishery industries worldwide. However, oysters are still at an early stage of domestication. One promising approach to solve the challenges is to use genome-editing techniques such as CRISPR/Cas9 system-mediated genomic editing technology.
Recently, the research team led by Prof. ZHANG Linlin from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) established a highly effective CRISPR-mediated gene editing technique in the cultured oyster, which also provides a powerful tool for genetic engineering breeding to improve productive traits in oysters and other aquaculture species.
Researchers performed CRISPR-mediated knockout by electroporation in Crassostrea gigas angulate with β-tubulin as a marker gene based on their previous established technics "mosaic mutations mediated by long fragment deletions". Through direct genotyping, long fragments deletions were detected in the target gene. By in situ hybridization, scanning electron microscopy, and behavioural analysis, mosaic mutations including defective cilia and decreased motility were observed in the G0 larva.
Compared with the small indels produced by single cleavages are too small to detect easily using normal agarose gels, the strategy of co-injecting more than two small guide RNAs (sgRNAs) was an important improvement to the protocol that significantly simplifies the genome editing workflow. According to genotyping, a long fragment deletion more than 300 bp was detected in the target gene.
Mosaicism resulting from CRISPR/Cas9 genome editing in animal models is generally regarded to be an undesirable outcome. However, in certain cases, this occurrence can be valuable. Because of the challenges posed by the embryonic lethality of many target genes, most of the attention has focused on analysis of mosaic G0 phenotypes. In this study, β-tubulin knockout mediated the mosaic expression patterns of Cgβ-tubulin and mosaic ciliary defects were commonly found at the positions of peritroch, intestinal cilia and the posterior cilium ring in C. gigas angulate larvae.
The sustainable development of the oyster genome editing breeding system calls for establishing efficient CRISPR/Cas9 mutagenesis tool that generating significant defective phenotypes. "In this study, we performed the CRISPR-mediated gene knockout by electroporation and described the long fragments deletions and mosaic mutations in the G0 larva of cultured oyster. The strategy significantly simplifies the genome editing workflow, and also provides an important new tool in the molluscs experimental system for studies of gene function," said Dr. CHAN Jiulin, first author of the study.
"The application of CRISPR-mediated gene editing technology in marine molluscs still faces great challenges, either in functional studies after gene knockout or during genetic engineering breeding. This study can provide useful reference for a widespread application of gene editing technology in the molluscs in the future," said Prof. ZHANG.
The above results were published in Frontiers in Marine Science on May 26, titled "Electroporation-based CRISPR/Cas9 Mosaic Mutagenesis of β-tubulin in the Cultured Oyster".
This study was supported by the Shandong Province Station "Techniques and Application of the Living Marine Resources" and Strategic Priority Research Program of the Chinese Academy of Sciences (B).
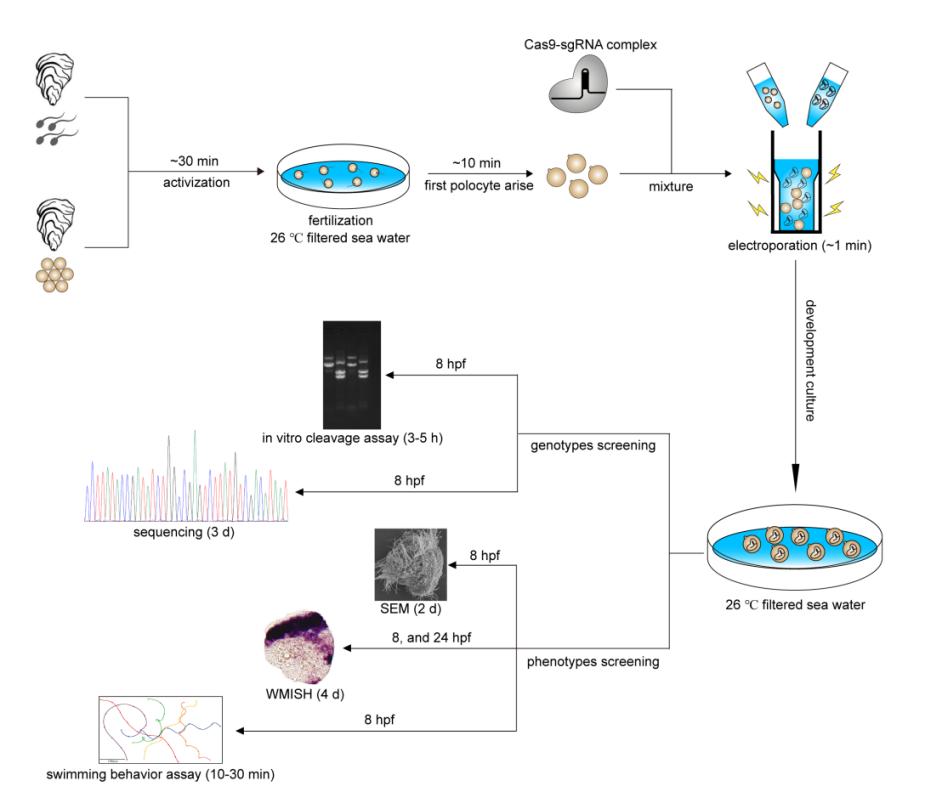
Procedure and timeline of G0 CRISPR/Cas9 mosaic knockout experiments in cultured oyster
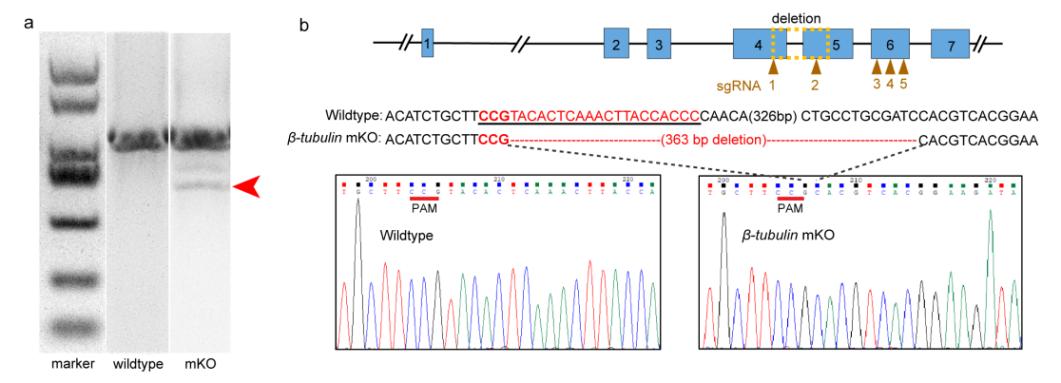
Characterization of deleted mutations in the electroporated embryos
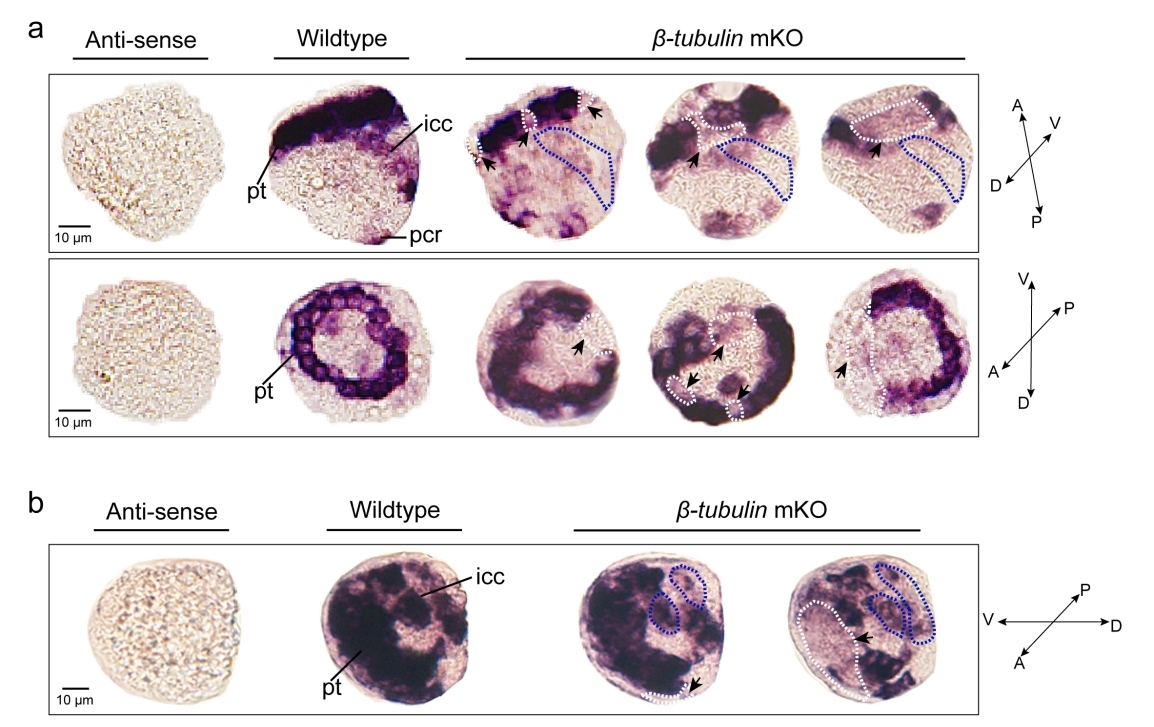
Effects of somatic mutagenesis in oyster larvae
Chan, J.#, Zhang, W.#, Xu, Y., Xue, Y., Zhang, L.* (2022). Electroporation-based CRISPR/Cas9 mosaic mutagenesis of β-tubulin in the cultured oyster. Frontiers in Marine Science, 9: 912409.
Zhang, L., Mazo-Vargas, A., Reed R.* (2017). A single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proceedings of the National Academy of the USA, 114(40):10707-12.
Zhang, L., Reed R.* (2016). Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nature Communications, 7: 11769.
ZHANG Linlin
Institute of Oceanology
E-mail: linlinzhang@qdio.ac.cn
(Editor: ZHANG Yiyi)

