The research team headed by Prof. Linsheng Song of IOCAS have made great process on the mechanism study of pufferfish heterosis, and the results were published by PLoS ONE on March 8, 2013.
This study was based on whole transcriptome analysis of Jiyan-1 hybrid pufferfish and its parental species Takifugu flavidus and Takifugu rubripes using SOLiD 4 NGS platform. The combined sequencing length was more than 12 Gbp, and a total of 44,305 transcripts were identified. From 14,148 differentially expressed transcripts, more than 2000 candidate genes and 35 candidate KEGG pathways of heterosis were determined. In addition, the co-existence of dominance, overdominance and additivity was proved. It indicated multiple gene action modes were involved in the heterosis of Jiyan-1. A bunch of novel alternative splicing isoforms with potential functions of ion binding, nucleic acid binding and kinase catalytic were identified in Jiyan-1, which might be related to the heterosis. This study provided valuable data sources for the better understanding of the determination and regulation of heterosis,and also was instructive in the application of heterosis.
This research was based on the powerful sequencing and computing capabilities of the marine bio-genetic resource platform and the supercomputing clusters in IOCAS. The publication of the results highlighted the innovation ability of the marine bio-genetic resources platform, and proved the adequate ability of conducting independent genomic/transcriptomic study with high-throughput sequencing technologies in IOCAS.
Full article can be accessed through:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0058453
This study was based on whole transcriptome analysis of Jiyan-1 hybrid pufferfish and its parental species Takifugu flavidus and Takifugu rubripes using SOLiD 4 NGS platform. The combined sequencing length was more than 12 Gbp, and a total of 44,305 transcripts were identified. From 14,148 differentially expressed transcripts, more than 2000 candidate genes and 35 candidate KEGG pathways of heterosis were determined. In addition, the co-existence of dominance, overdominance and additivity was proved. It indicated multiple gene action modes were involved in the heterosis of Jiyan-1. A bunch of novel alternative splicing isoforms with potential functions of ion binding, nucleic acid binding and kinase catalytic were identified in Jiyan-1, which might be related to the heterosis. This study provided valuable data sources for the better understanding of the determination and regulation of heterosis,and also was instructive in the application of heterosis.
This research was based on the powerful sequencing and computing capabilities of the marine bio-genetic resource platform and the supercomputing clusters in IOCAS. The publication of the results highlighted the innovation ability of the marine bio-genetic resources platform, and proved the adequate ability of conducting independent genomic/transcriptomic study with high-throughput sequencing technologies in IOCAS.
Full article can be accessed through:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0058453
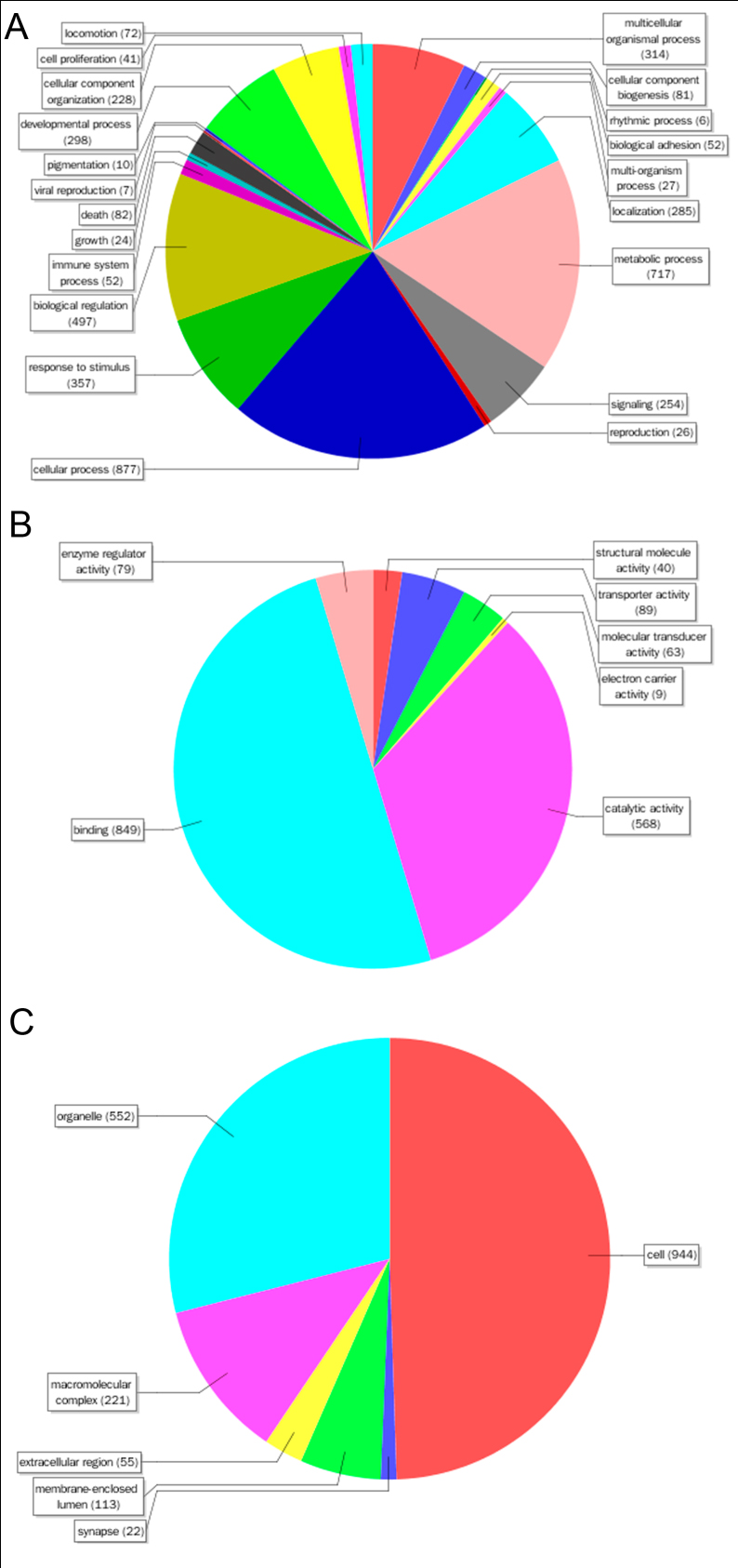
Figure. Sequence distribution by GO for heterosis candidate genes.
A. biological process. B. molecular function.
C. cellular component. Numbers shown in the chart represented the amount of sequences with corresponding GO terms.

