As foundational species in coastal ecosystems, only about 70 seagrass species have been documented worldwide—a surprisingly low number given their global distribution and ecological significance. Moreover, seagrass phenotypes are highly plastic within species, which further complicates species-level determination.
Recently, the research team led by Prof. ZHOU Yi from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS), together with their collaborators from GEOMAR Helmholtz Centre for Ocean Research Kiel in Germany and other institutions, have discovered one cryptic speciation event within Nanozostera japonica, a seagrass species widely distributed across the Northwest Pacific.
The study was published in New phytologist on Jun. 30.
N. japonica is one of the few seagrass species distributed across both temperate and tropical-subtropical coastal zones. It is native in the Northwest Pacific, and spread to the North American Pacific coast in the early 20th century via oyster transportation. N. japonica exhibits highly variable phenotypes among different geographic environments. Moreover, the researchers used microsatellite markers to reveal pronounced genetic differentiation between northern and southern populations. Thus, they proposed that the contemporary N. japonica populations may contain different species.
To test this hypothesis, they first assembled two high-quality, chromosome-level reference genomes using N. japonica samples collected from the northern and southern China, respectively. Then they conducted whole-genome resequencing of 17 geographic populations across the Western Pacific. Genomic analysis showed that the divergence between the northern and southern genetic clades happened ca. 4.16 Ma—surprisingly, the southern clade was more closely related to its European sister species N. noltii, with a more recent divergence of just 2.67 Ma.
"The genetic divergence between the two N. japonica clades exceeds typical intro-species difference," said Associate Prof. ZHANG Xiaomei.
The study also identified a few hybrids between the two clades in the contact zone, all of which were exclusively first-generation (F1) diploids or triploids, without higher order hybrids. This pattern strongly suggests reproductive isolation between the two putative species.
Further comparative genomic analyses revealed a massive ~42 Mb chromosomal inversion showing fixed differences between the two clades, which might have contributed to the reproductive separation.
"This work demonstrates that what we currently recognize as N. japonica actually represents two distinct species, providing critical insights for future seagrass classification and conservation strategies," said Prof. ZHOU.
This is the first time that cryptic species in seagrasses were identified going full population genomics in seagrasses. This study suggest that seagrass diversity may be underestimated, highlighting the need for more comprehensive population genomic studies in this ecologically vital group.
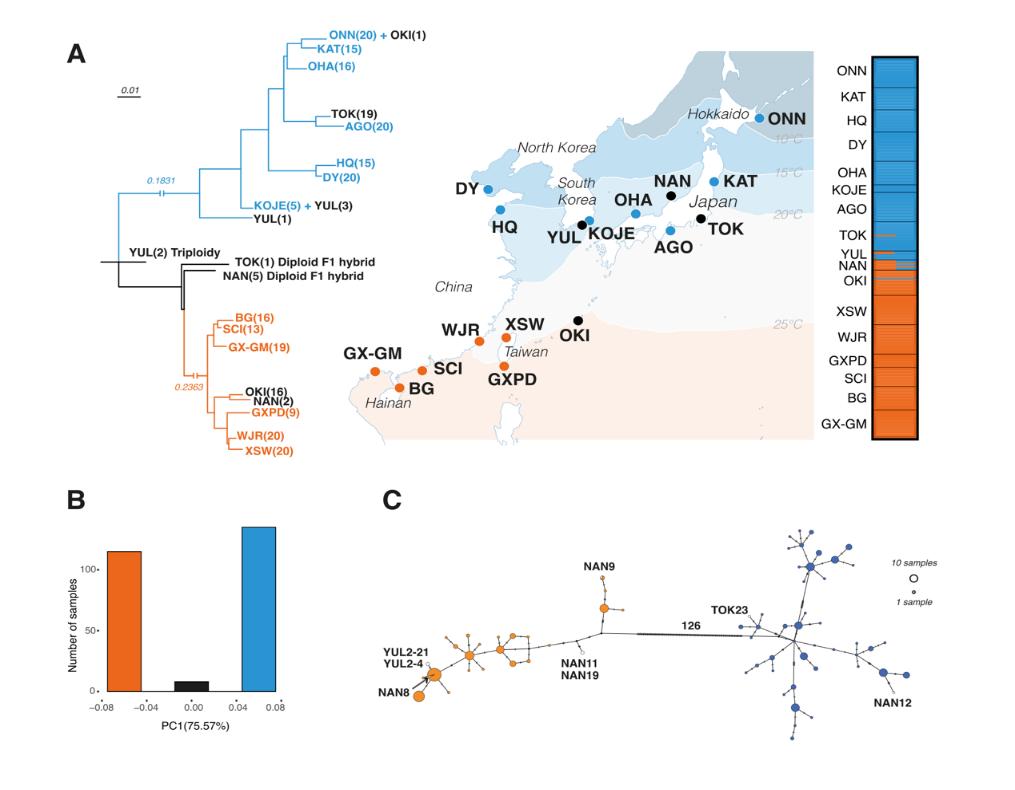
Fig. 1 Genetic population structure of the Nanozostera japonica species complex. (Image by IOCAS)
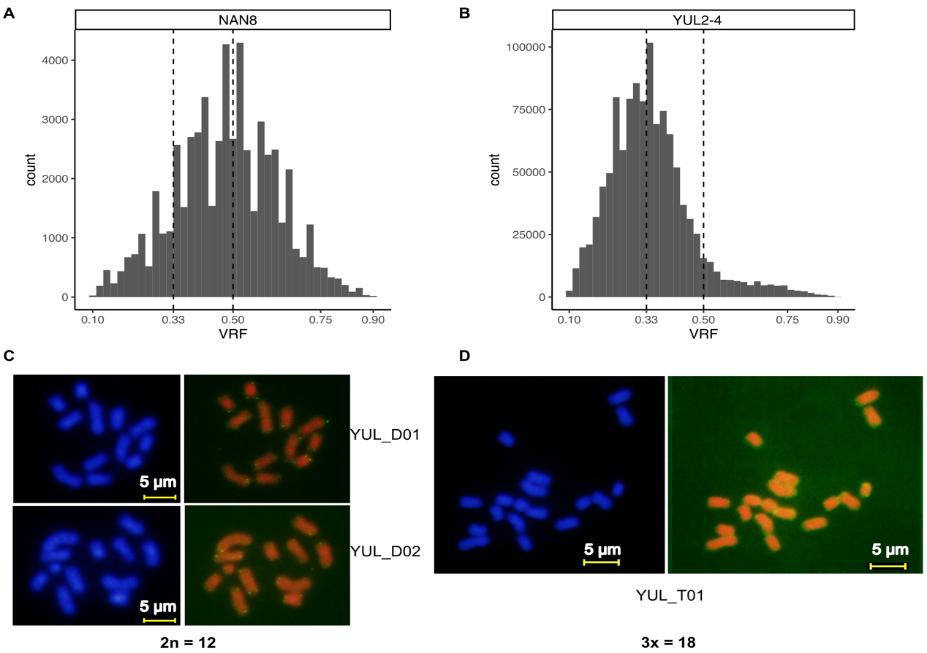
Fig. 2 Co-existence of diploidy and triploidy within a population of Nanozostera japonica. (Image by IOCAS)
(Text by Prof. ZHANG Xiaomei)
Media Contact:
ZHANG Yiyi
Institute of Oceanology
E-mail: zhangyiyi@qdio.ac.cn
(Editor: ZHANG Yiyi)

