Understanding the genetic mechanisms of adaptive evolution represents a fundamental challenge in molecular biology. While organisms can rapidly adapt phenotypically to environmental changes, the underlying genomic processes remain poorly understood.
The transition of fish from marine to freshwater environments is a complex adaptive process involving multiple polygenic traits, making the parallel evolution system of rapid freshwater adaptation an ideal model for deciphering the genetic architecture underlying phenotypic adaptation to novel environments.
Recently, the research team led by Prof. LIU Jin-Xian from the Institute of Oceanology Chinese Academy of Sciences (IOCAS) investigated the genetic mechanisms underlying the rapid freshwater adaptation of a Salangid icefish (Neosalanx brevirostris) by comparing genomic differences between the ancestral anadromous population and the derived freshwater-resident populations.
The study was published in Molecular Biology and Evolution on Jul. 03.
The researchers revealed that distinct freshwater-resident populations within the Yangtze River basin derived independently from the Yangtze River Estuary population, thus forming a parallel freshwater adaptation system. Further analysis confirmed that rapid parallel freshwater adaptation follows a complex polygenic architecture and demonstrated parallelism at the genome level, which was predominantly driven by selection on standing genetic variants.
In the ancestral Yangtze River Estuary population, the frequency of candidate adaptive genetic variations was at a moderate level, suggesting that this population had already undergone pre-adaptation to low-salinity conditions in the fluctuating estuarine environment, significantly accelerating the pace of parallel freshwater adaptation.
During parallel freshwater adaptation, relatively large allele frequency shifts were observed in the adaptive SNPs, with a large fraction of freshwater favored alleles becoming either fixed or nearly fixed. These adaptive SNPs are functionally associated with biological processes such as osmoregulation, immunoregulation, locomotion, and metabolism. "This was consistent with the polygenic architecture underlying adaptive divergence between the two ecotypes, which involves complex physiological and behavioral traits," said LI Yulong, the study's first author.
This case study highlights the critical role of standing genetic variation in the adaptive evolution of populations undergoing environmental changes and offers new insights for further investigation into the molecular mechanisms underlying rapid adaptive evolution of complex biological traits.
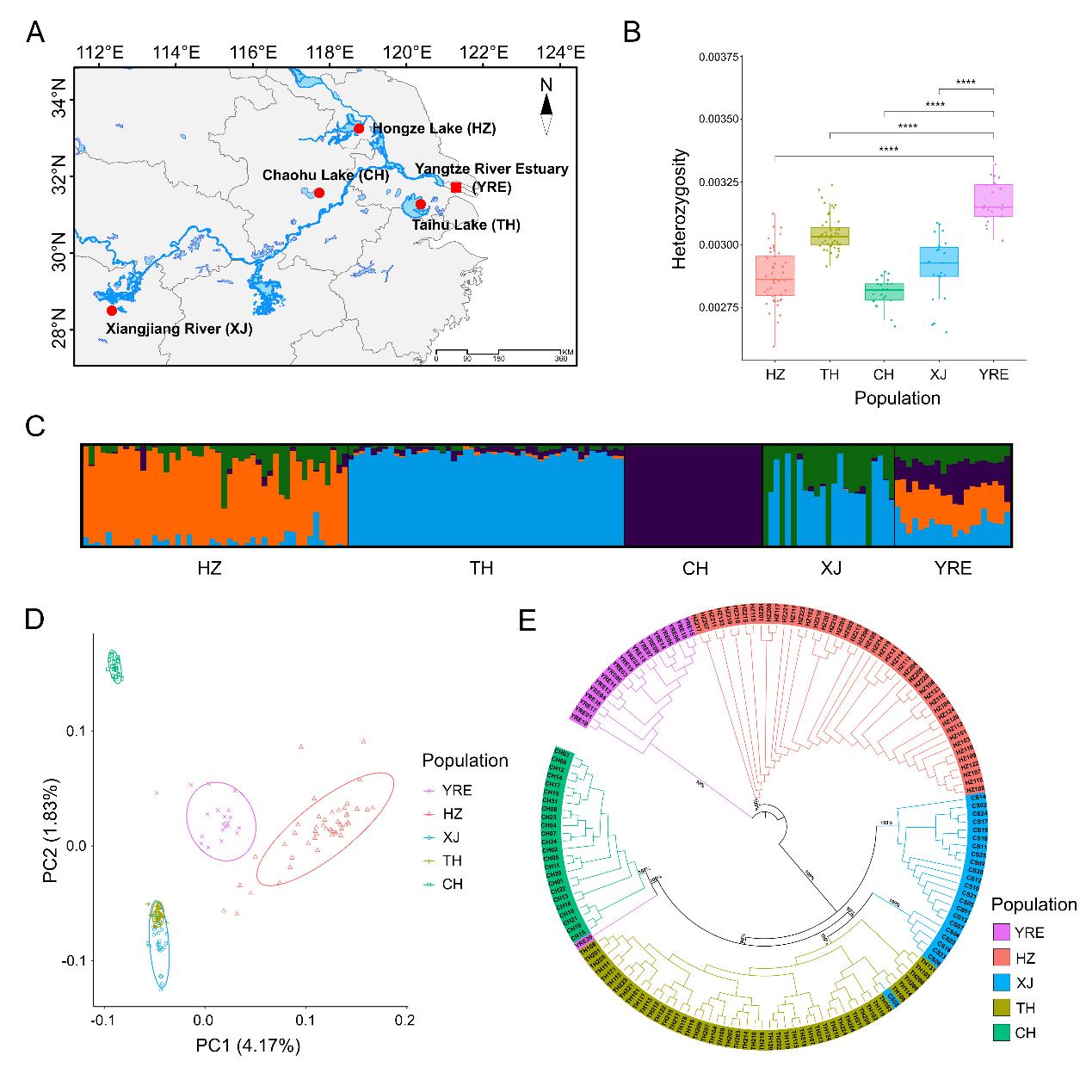
Sampling map, genetic diversity, and population genetic structure of Neosalanx brevirostris populations. (Image by IOCAS)
(Text by YANG Hao)
Media Contact:
ZHANG Yiyi
Institute of Oceanology
E-mail: zhangyiyi@qdio.ac.cn
(Editor: ZHANG Yiyi)

