By deciphering the whole genome sequence of a scallop, researches revealed a slowly evolving genome and proposed a novel expression pattern of Hox genes, the subcluster temporal co-linearity (STC). Recently, these findings were published in Nature Ecology & Evolution entitled “Scallop genome provides insights into evolution of bilaterian karyotype and development” (DOI: 10.1038/s41559-017-0120).
The genomes of marine animals contain crucial information to understand animal evolution because all animal phyla emerged in the ocean and most of them still only inhabit there. In the abovementioned study, researchers from China, USA and Norway studied the whole genome of the Yesso scallop Patinopecten yessoensis, which lives in relatively cold water. Chromosome-based macrosynteny analysis revealed a striking correspondence between the 19 scallop chromosomes and the 17 presumed ancestral bilaterian linkage groups at a level of conservation previously unseen, suggesting that the scallop may have a karyotype close to that of the bilaterian ancestor. In addition, by investigating the expression of Hox genes, researchers proposed a novel expression pattern of Hox genes, the subcluster temporal co-linearity (STC).
Liu and Huan from IOCAS participated in the project. They studied the spatiotemporal expression of all 11 scallop Hox genes during development and revealed the spatial co-linearity expression of the “leading” Hox genes. These results well support the STC hypothesis and thus provide essentialevidenceto the study.
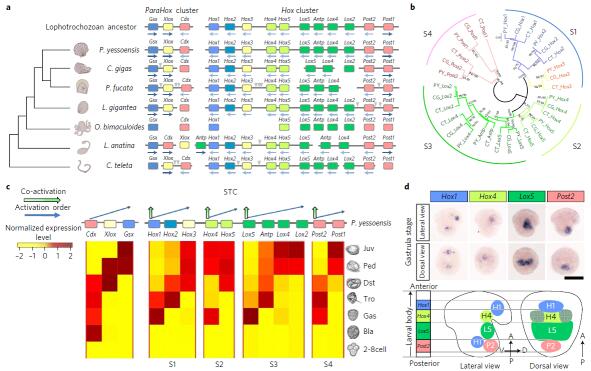
Fig. a: Organization of Hox genes in P. yessoensis; b: Phylogenetic analysis between P. yessoensis Hox genes and other lophotrochozoan; c: Transcriptomic data support subcluster temporal co-linearity expression of Hox genes; d: Expression of the four “leading” Hox genes exhibits spatial co-linearity in gastrulae.
Contact: Dr. Huan Pin, Institute of Oceanology
Email:huanpin@qdio.ac.cn
|
|

Address: 7 Nanhai Road, Qingdao, Shandong 266071, China
Tel: 86-532-82898902 Fax: 86-532-82898612 E-mail: iocas@qdio.ac.cn


