Unusual structural features of the dUTPase of white spot syndrome virus provide a unique opportunity for antiviral development
White spot syndrome virus (WSSV) is a serious pathogen in shrimp aquaculture, causing $1 billion economic loss every year since its emergence more than 20 years ago. It is a large DNA virus and the sole member of the genus Whispovirus and the family Nimaviridae. So far, its replication mechanism is not understood and no effective treatment is available. WSSV genome encodes more than 180 proteins and most of them have unclear functions. Interestingly, a rather complete thymidylate biosynthesis pathway is encoded in WSSV genome, despite the host can provide thymidylate. dUTPase is an enzyme in this pathway, essential for both thymidylate synthesis and viral genome integrity maintenance. Recently, researchers from the Institute of Oceanology, Chinese Academy of Science have found that WSSV dUTPase adopts a structural form distinct from classic dUTPases, providing a unique opportunity to develop antiviral drugs against WSSV infections.
dUTPase catalyzes the hydrolysis of dUTP to dUMP and pyrophosphate (PPi), playing essential roles in cellular nucleotide metabolism. Interestingly, the trimeric dUTPase of WSSV (wDUT) harbors a sequence insert at the position preceding the C-terminal catalytic motif V (pre-V insert), rarely seen in other dUTPases. Through X-ray crystallographic analysis combined with enzymatic assay, we observed that the pre-V insert in wDUT forms an unusual β-hairpin structure in the domain-swapping region, and thereby facilitates a unique orientation of the adjacent C-terminal segment, positioning the catalytic motif V onto the active site of its own subunit instead of a third subunit. Consequently, wDUT employs two-subunit active sites, unlike the widely accepted paradigm that the active site of trimeric dUTPase is contributed by all three subunits. According to results from local structural comparisons, the active-site configuration of wDUT is similar to that of known dUTPases. However, we also found that residues in the second-shell region of the active site are reconfigured in wDUT as an adaption to its unique C-terminal orientation. We hypothesize that this rare structural arrangement confers additional functionality to wDUT. In summary, our study discovers a novel structural form of dUTPase and demonstrates how sequence insert and amino acid substitution in concert drive protein evolution. The structural information is valuable for antiviral drug design against the white spot syndrome virus.
The finding has been published in Journal of Biological Chemistry entitled “The dUTPase of white spot syndrome virus assembles its active sites in a noncanonical manner” (http://www.jbc.org/content/293/3/1088). The work was supported by the National Natural Science Foundation of China, the “1000 Talents Program” of China, the “100 Talents Program” of the Chinese Academy of Sciences, and the “AoShan Talents Program” of Qingdao National Laboratory for Marine Science and technology.
Corresponding author: Qingjun Ma (qma@qdio.ac.cn), Institute of Oceanology, CAS
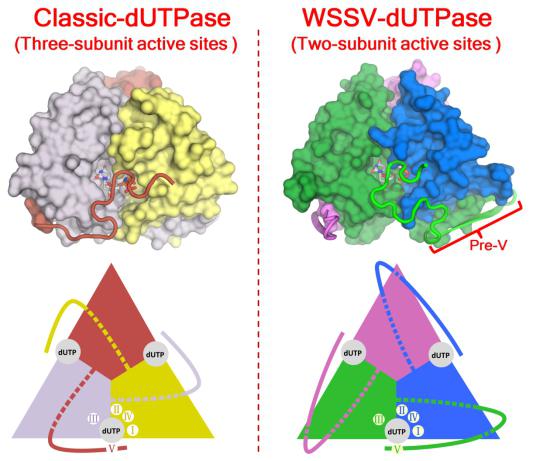
Comparison of active site assembly between classic dUTPase and WSSV dUTPase




