“Divergence and plasticity shape adaptive potential of the Pacific oyster”, this 5 years study is a follow-up of another major achievement published in the Nature in 2012 for Prof. Guofan Zhang’s research group in the field of marine organism adaptation.
Due to the global climate change and inappropriate human activities, marine organisms are facing varying degrees of environmental challenges. Its adaptive potential and mechanism are the focus in the field of ocean and evolution. With ecological and economic importance, the oyster is the earliest and the most studied mollusk. Oysters and intertidal zone provide an excellent system for the study of marine organism adaptive evolution.

Fig. 1 Wild oyster inhabiting intertidal zone at Qingdao of China
The fine-scale genetic and phenotypic divergence across environmental gradients suggest that selection and local adaptation are pervasive and, together with limited gene flow, influence population structure of the wild population of China. Genes showing sequence differentiation between populations also diverged in transcriptional response to heat stress. Plasticity in gene expression is positively correlated with evolved divergence, indicating that plasticity is adaptive and favored by organisms under dynamic environments. Divergence in heat tolerance—partly through acetylation-mediated energy depression—implies differentiation in adaptive potential. Trade-offs between growth and survival may play an important role in local adaptation of oysters and other marine invertebrates.


Fig. 2 Team members are collecting the wild oysters and doing spawning in the hatchery
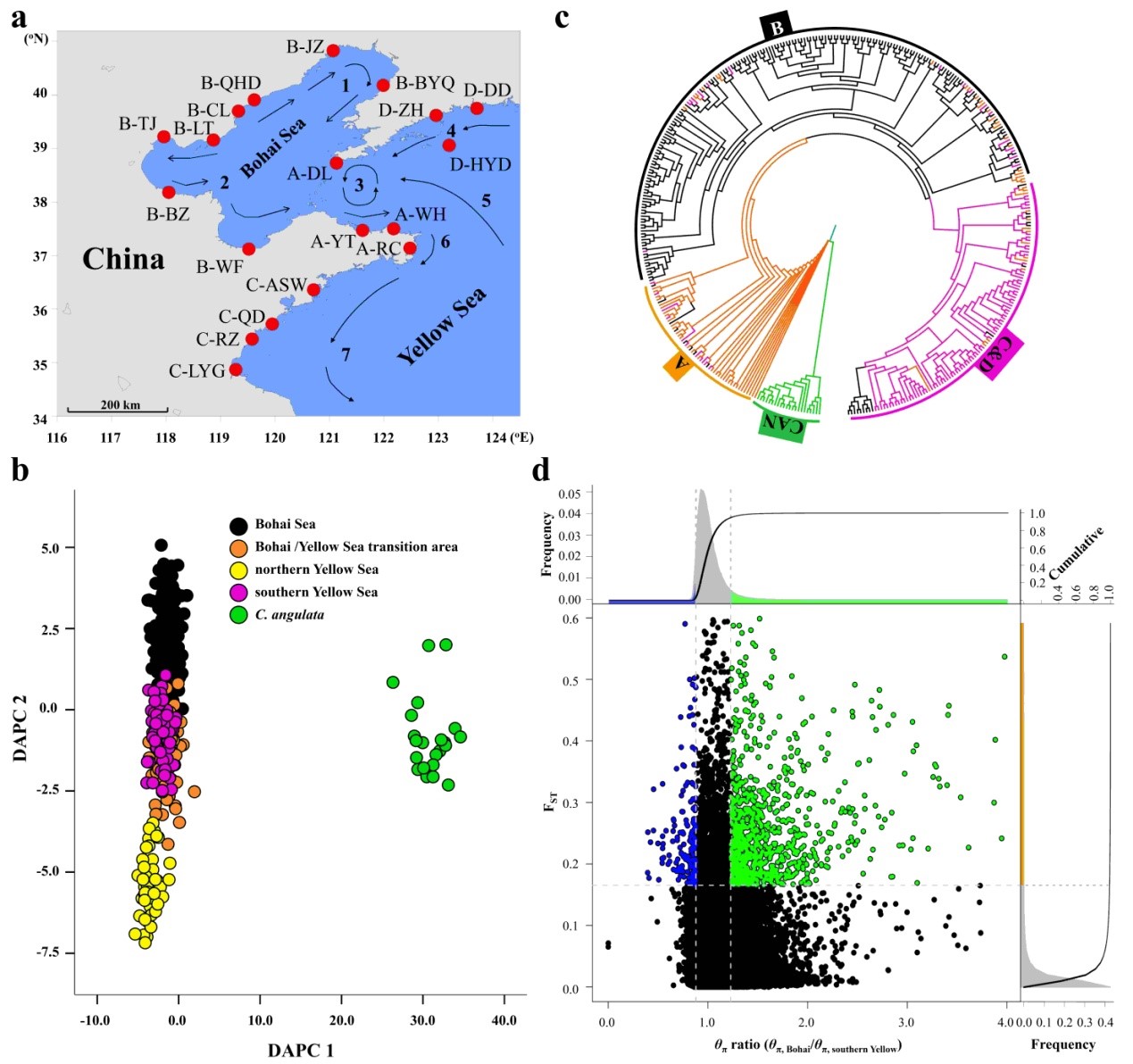
Fig. 3 Geographic distribution and population divergence of Pacific oysters in northern China. a, Collection sites of re-sequenced Pacific oysters from northern China. b, Scatterplot of DAPC inferred from whole-genome SNPs. c, Phylogenetic tree of Pacific oysters inferred from whole-genome SNPs by the neighbour-joining method, using C. angulata (CAN) as the outgroup. d, Distribution of θπ ratios (θπ,BS/θπ,SYS) and FST values calculated in 10-kb windows sliding in 2-kb steps.
These findings provide insights into how marine populations adapt and diverge under complex conditions, and contribute to the understanding of the evolutionary potential of marine organisms under rapid climate change, as well as evaluation of genetic resources for conservation and sustainable ecosystem services.
Please find the original paper at: https://www.nature.com/articles/s 41559-018-06 68-2
|
|

Address: 7 Nanhai Road, Qingdao, Shandong 266071, China
Tel: 86-532-82898902 Fax: 86-532-82898612 E-mail: iocas@qdio.ac.cn


