Interactions between algae and bacteria represent an important inter-organismal association in aquatic environments, which often have cascading bottom-up influences on ecosystem-scale processes. This has become a promising research theme in understanding the ecology of both algae and bacteria.
Despite increasing recognition of linkages between bacterioplankton and dynamics of dinoflagellate blooms in the field, the knowledge about the forms and functions of dinoflagellate-bacterium associations remains elusive, mainly due to the ephemeral and variable conditions in the field.
Recently, the research team led by Prof. TANG Ying-Zhong from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) provided new insights into the fundamental functions of bacteria consortia associated with the phycospheres of dinoflagellates and other harmful algal blooms (HABs)-forming microalgae.
The study was published in International Journal of Environmental Research and Public Health on Apr. 7.
Researchers characterized the bacterial assemblages associated with 144 clonal cultures of harmful algae that have been established and cultured in the laboratory, including 130 strains of dinoflagellates (covering all major taxa of dinoflagellates) and 14 strains from other classes, via high-throughput sequencing for 16S rRNA gene amplicons.
The co-occurrence of highly diversified bacteria implies that the association between these algal species and bacterial consortia is either bilaterally (i.e., mutualism) or at least unilaterally (i.e., commensalism) beneficial to the two partners.
Bacterial communities in dinoflagellates cultures exhibited remarkable conservation across different algal strains, which were dominated by a relatively small number of taxa, most notably the γ-proteobacteria Methylophaga, Marinobacter and Alteromonas.
Although the bacterial community composition did not show significant difference between dinoflagellates and non-dinoflagellate groups, dinoflagellates appeared to harbor a larger number of the "unique features" (equivalent to species in the context) that had relatively low abundance and to be enriched in Methylophaga, supposedly being methylotroph.
"While the bacterial assemblages associated with thecate and athecate dinoflagellates displayed no general difference in species composition and functional groups, athecate dinoflagellates appeared to accommodate more aerobic cellulolytic members of Actinobacteria, implying a more possible reliance on cellulose utilization as energy source," said Dr. DENG Yunyan, first author of the study.
"We believe our results provided insightful understanding of the species composition and community functional profiles of dinoflagellate-associated bacterial assemblages, and the work has also paved step stones for some more interesting and significant investigations," said Prof. TANG.
The research was supported by the National Science Foundation of China, the Key Deployment Project of Centre for Ocean Mega-Research of Science, Chinese Academy of Sciences and the Natural Science Foundation of Shenzhen, China.
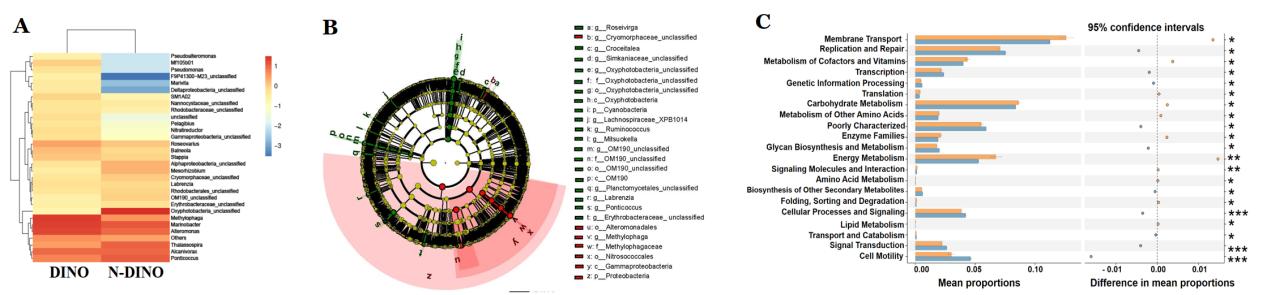
Heatmap (A), phylogenetic distribution (B) and differential function prediction of bacterial communities differentially represented between dinoflagellates and non-dinoflagellates
Deng, Y. Y., Wang, K., Hu, Z. X., and Tang, Y. Z. (2022). Abundant Species Diversity and Essential Functions of Bacterial Communities Associated with Dinoflagellates as Revealed from Metabarcoding Sequencing for Laboratory-Raised Clonal Cultures. International Journal of Environmental Research and Public Health, 19: 4446.
TANG Ying-Zhong
Institute of Oceanology
E-mail: yingzhong.tang@qdio.ac.cn
(Editor: ZHANG Yiyi)
|
|

Address: 7 Nanhai Road, Qingdao, Shandong 266071, China
Tel: 86-532-82898902 Fax: 86-532-82898612 E-mail: iocas@qdio.ac.cn


